G3:中科院东北地理所冯献忠研究组发表大豆基因遗传定位方法研
10月19日 《G3:GENEs|Genomes|genetics》杂志上在线发表中国科学院东北地理所冯献忠研究组发表的大豆基因遗传定位方法研究成果。东北地理所大豆分子设计育种重点实验室的合作培养博士生宋晓峰和该所博士魏海超为论文共同第一作者,研究员冯献忠和教授张慧为论文责任作者。
由于大豆基因组大和基因组复杂,大豆重要农艺性状基因的定位克隆一直是大豆分子遗传学研究中的难点。中国科学院东北地理与农业生态研究所大豆功能基因组学学科组的研究人员通过与山东师范大学、中科院上海生命科学研究院和中山大学等科研单位合作,建立了利用全基因组测序信息开发INDEL分子标记快速定位大豆基因的新方法。
研究组成员通过对大豆“菏豆12”品种的全基因组重测序,获得了与“Willams82”品种存在差异的49,276个INDEL分子标记和242,059个SNP分子标记。通过对其中243个INDEL分子标记的实验验证,建立了165个INDEL分子标记大豆分子标记物理图谱。利用该图谱和进一步开发的INDEL分子标记成功地将大豆叶皱突变体定位到了360Kbp的关联区间。该研究为大豆重要农艺性状基因克隆和分子辅助育种等研究提供了重要的研究手段。
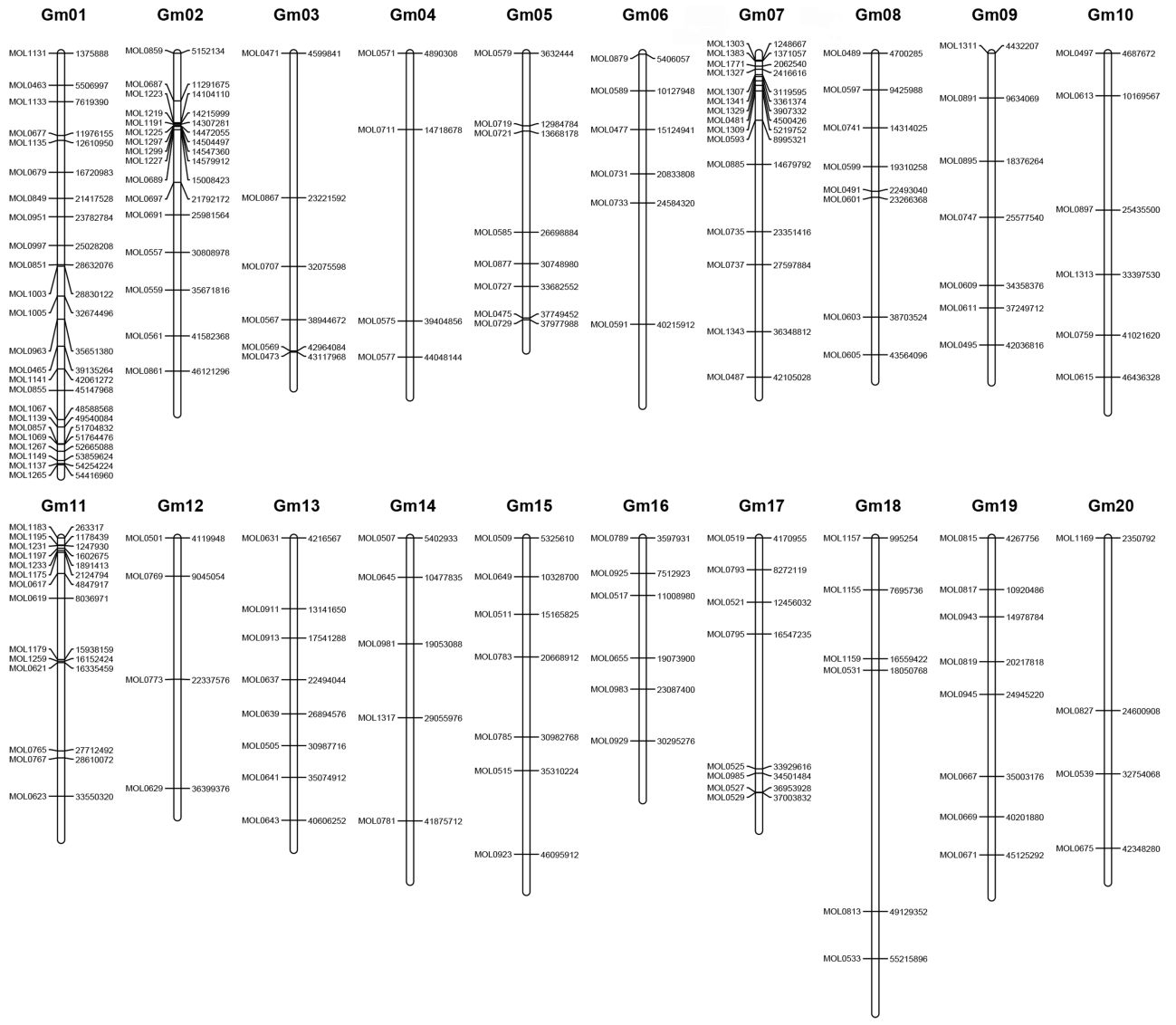
大豆INDEL分子标记物理图谱
原文链接:
Development of INDEL markers for Genetic Mapping Based on Whole-Genome Re-sequencing in Soybean
原文摘要:
Soybean (Glycine max (L.) Merrill) is an important crop in the world. In this study, a Chinese local soybean cultivar, Hedou 12, was re-sequenced by next generation sequencing technology to develop INsertion/DELetion (INDEL) markers for genetic mapping. 49,276 INDEL polymorphisms and 242,059 single nucleotide polymorphisms (SNP) were detected between Hedou 12 and the Williams 82 reference sequence. Of these, 243 candidate INDEL markers ranging from 5 to 50 bp in length were chosen for PCR validation, and 165 (68%) of them revealed polymorphisms between Hedou 12 and Williams 82. The validated INDEL markers were also tested in 12 other soybean cultivars. The number of polymorphisms in the pairwise comparisons of 14 soybean cultivars varied from 27 to 165. To test the utility of these INDEL markers, they were used to perform a genetic mapping of a crinkly leaf mutant, and the CRINKLY LEAF locus was successfully mapped to a 360 Kb region on chromosome 7. This research shows that high-throughput sequencing technologies can facilitate the development of genome-wide molecular markers for genetic mapping in soybean.
作者:冯献忠

