中科院田朝光组全面解析原绿球藻MED4的遗传子图谱
中科院天津工业生物技术研究所田朝光研究组以原绿球藻的典型代表——MED4为例,利用第二代测序技术(RNA-Seq),全面解析了原绿球藻MED4的操纵子图谱,转录本非翻译区以及新基因。相关文章发表于2014年1月18日的《BMC Microbiology》杂志上。
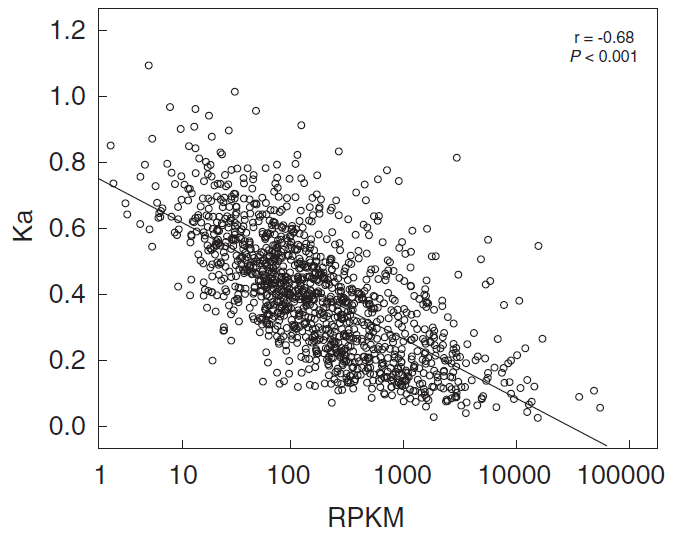
图1 基因表达水平与非同义突变之间的负相关关系
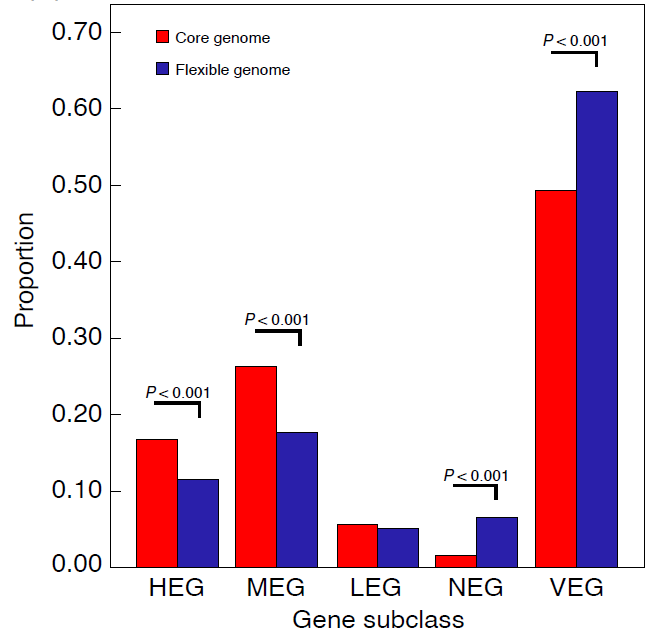
图2 核心基因组与非核心基因组各表达水平基因所占比例
原绿球藻(Prochlorococcus)是分布在全球热带和亚热带海域的蓝细菌,也是目前发现的海洋中含量最丰富的光合生物。虽然只具有极小的细胞体积(直径约0.6 um)和基因组(可以小到~1.6Mbp),但原绿球藻细胞数量的庞大以及高效的光合作用,使得其对于全球生物圈初级产物的生产和碳循环具有极其重要的地位。另一方面,目前已有十多种原绿球藻生态亚种的基因组被发布,亚种之间基因组丢失的现象很早就被人们发现并探讨过。
但是,在这种基因得失剧烈的背景下,仍然有约1250个蛋白编码基因为目前测序的原绿球藻所共有,其构成了原绿球藻的核心基因组(Core genome)。探索稳定蓝细菌核心基因组的力量,不仅是进化生物学的重要命题,同时对于更深入地理解最小基因组的内涵和外延,乃至运用简约基因组的概念指导光合系统合成细胞的开发,都具有重要意义。
中科院天津工业生物技术研究所田朝光研究组以原绿球藻的典型代表——MED4为例,利用第二代测序技术(rna-seq),全面解析了原绿球藻MED4的操纵子图谱,转录本非翻译区以及新基因。
基于全基因组的表达数据,发现基因表达水平(RPKM)与分子进化速率(Ka)之间存在显著的负相关关系(N=1275,Spearman’s r=-0.68,P < 0.001;如图1)。通过进一步比较MED4核心基因和非核心基因在基因必要性和基因表达水平上的差异,发现两者能相对独立地影响分子进化速率;即基因表达水平越高,基因越重要,进化越慢。
由于核心基因组比非核心基因组(Flexible genome)拥有更高的表达水平 (图2)和不可丢失性,所以核心基因组在原绿球藻数亿甚至几十亿年的进化中得以保留下来。此外,高表达的mRNA更加快速的降解,能提高核心基因组蛋白翻译的鲁棒性(Robustness),进而减少细胞毒性和能量浪费。
该研究得到国家973计划等资助。中科院硕士研究生王邦为论文第一作者。
原文摘要:
The transcriptome landscape of Prochlorococcus MED4 and the factors for stabilizing the core genome
Bang Wang, Lina Lu, Hexin Lv, Huifeng Jiang, Ge Qu, Chaoguang Tian and Yanhe Ma
Background
Gene gain and loss frequently occurs in the cyanobacterium Prochlorococcus, a phototroph that numerically dominates tropical and subtropical open oceans. However, little is known about the stabilization of its core genome, which contains approximately 1250 genes, in the context of genome streamlining. Using Prochlorococcus MED4 as a model organism, we investigated the constraints on core genome stabilization using transcriptome profiling.
Results
RNA-Seq technique was used to obtain the transcriptome map of Prochlorococcus MED4, including operons, untranslated regions, non-coding RNAs, and novel genes. Genome-wide expression profiles revealed that three factors contribute to core genome stabilization. First, a negative correlation between gene expression levels and protein evolutionary rates was observed. Highly expressed genes were overrepresented in the core genome but not in the flexible genome. Gene necessity was determined as a second powerful constraint on genome evolution through functional enrichment analysis. Third, quick mRNA turnover may increase corresponding proteins’ fidelity among genes that were abundantly expressed. Together, these factors influence core genome stabilization during MED4 genome evolution.
Conclusions
Gene expression, gene necessity, and mRNA turnover contribute to core genome maintenance during cyanobacterium Prochlorococcus genus evolution.
Keywords:
Core genome; Gene expression; Molecular evolution; Prochlorococcus; RNA-Seq; Transcriptome

