GBE:中科院李德铢等运用叶绿体系统发育基因组学明确岩菖蒲科是
2016年3月8日,国际著名学术期刊《Genome Biology and evolution》在线发表了中国科学院昆明植物研究所李德铢研究组和王红研究组的博士生骆洋与马朋飞博士等人合作的“Plastid phylogenomic analyses resolve Tofieldiaceae as the root of the early diverging monocot order Alismatales”。该研究对于揭示单子叶植物分支早期的起源和演化具有重要的科学意义,也为利用叶绿体系统发育基因组学确定被子植物演化的关键节点提供了一个重要案例。
泽泻目(Alismatales)是单子叶植物分支(Monocots)中的一个非常重要的水生类群,并且可能是被子植物最早“下海”的一个祖传系,在淡水和海岸带生态系统中扮演着重要的角色。在当前通行的APG系统中,泽泻目是单子叶植物在其最基部的菖蒲目(Acorales)后随即分化出来的基部类群。十余年来,基于dna片段所做的分子系统学研究似乎支持天南星科(Araceae)为泽泻目的基部类群,但仍存在诸多疑问。
基于叶绿体系统发育基因组学探讨了单子叶植物的早期演化,该团队首次测定了岩菖蒲科(Tofieldiaceae)、眼子菜科(Potamogetonaceae)和泽泻科(Alismataceae)几个代表种的叶绿体全基因组,发现利川慈姑(Sagittaria lichuanensis)的叶绿体基因组总长为179,007bp,是目前在单子叶植物中发现的最大的叶绿体基因组,并在慈姑属(Sagittaria)和眼子菜属(Potamogeton)中分别发现了一个约2.4kb和6kb的倒位。选取79个叶绿体蛋白质编码基因,基于不同的数据分析方法重建了早期分化的单子叶植物分支的分子系统发育框架,明确了泽泻目为单系,岩菖蒲科是泽泻目最基部的类群。
该研究得到了国家重大科学研究计划项目(2014CB954100)、国家自然科学基金委重大国际合作项目(31320103919)、国家自然科学基金面上项目(31270272)和中国科学院“青年创新促进会”人才专项(2015321)的支持。
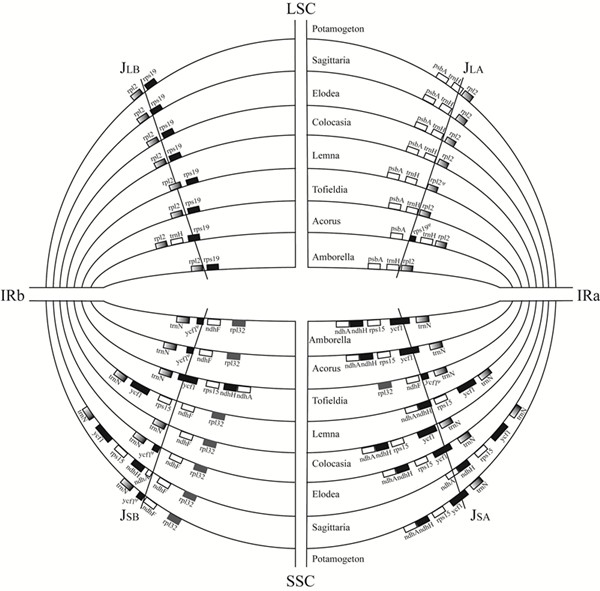
图1 单子叶植物分支早期分化类群的叶绿体基因组结构对比
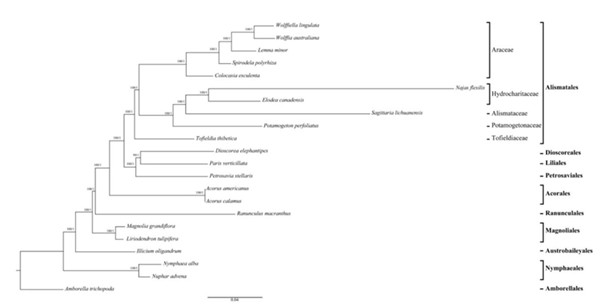
图2 基于79个蛋白质编码基因的22个类群的系统发育树
原文链接:
原文摘要:
The predominantly aquatic order Alismatales, which includes approximately 4500 species within Araceae, Tofieldiaceae, and the core alismatid families, is a key group in investigating the origin and early diversification of monocots. Despite their importance, phylogenetic ambiguity regarding the root of the Alismatales tree precludes answering questions about the early evolution of the order. Here, we sequenced the first complete plastid genomes from three key families in this order:Potamogeton perfoliatus (Potamogetonaceae), Sagittaria lichuanensis(Alismataceae), and Tofieldia thibetica (Tofieldiaceae). Each family possesses the typical quadripartite structure, with plastid genome sizes of 156,226 bp, 179,007 bp, and 155,512 bp, respectively. Among them, the plastid genome of S. lichuanensis is the largest in monocots and the second largest in angiosperms. Like other sequenced Alismatales plastid genomes, all three families generally encode the same 113 genes with similar structure and arrangement. However, we detected 2.4 kb and 6 kb inversions in the plastid genomes of Sagittaria and Potamogeton, respectively. Further, we assembled a 79 plastid protein-coding gene sequence data matrix of 22 taxa that included the three newly generated plastid genomes plus nineteen previously reported ones, which together represent all primary lineages of monocots and outgroups. In plastid phylogenomic analyses using maximum likelihood and Bayesian inference, we show both strong support for Acorales as sister to the remaining monocots and monophyly of Alismatales. More importantly, Tofieldiaceae was resolved as the most basal lineage within Alismatales. These results provide new insights into the evolution of Alismatales as well as the early-diverging monocots as a whole.
作者:骆洋

