Nuleic Acids Res:中科院上海细胞所王恩多研究组揭示细菌来源的
7月21日,国际学术期刊《核酸研究》(Nucleic Acids Research)在线发表了中国科学院上海生命科学研究院生物化学与细胞生物学研究所王恩多研究组题为“tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily”的最新研究成果。
tRNA作为连接遗传信息和对应氨基酸之间的“接头分子”,是蛋白质合成中的关键生物大分子之一。tRNA的核苷酸残基上存在着广泛的转录后修饰,这些修饰对于tRNA在细胞内发挥功能起着重要作用。其中tRNA第32位核苷酸核糖上的2'-O-甲基化修饰在所有生物中存在。SPOUT甲基化酶超家族成员TrmJ是存在于细菌以及古菌中的tRNA 32位2'-O-甲基化酶。与古菌TrmJ不同,细菌来源的TrmJ需要识别全长的tRNA作为底物。但是,TrmJ如何识别tRNA和催化tRNA的甲基化的机制仍不清楚。
王恩多研究组刘如娟副研究员、博士研究生龙韬等人通过解析2.6Å分辨率的大肠杆菌的TrmJ(EcTrmJ)与辅因子S-腺苷-高半胱氨酸(SAH)复合物的晶体结构以及定点突变、等温滴定量热(ITC)实验、凝胶迁移实验(EMSA)及酶学动力学等生物化学手段研究了EcTrmJ与tRNA底物的结合方式。研究结果表明,EcTrmJ形成不对称的同源二聚体,其SPOUT结构域和延伸结构域均能独立形成二聚体;而这两个结构域都参与对底物tRNA的识别,连接两体的一段不保守柔性铰链区在与底物的识别过程中起关键的调节作用。
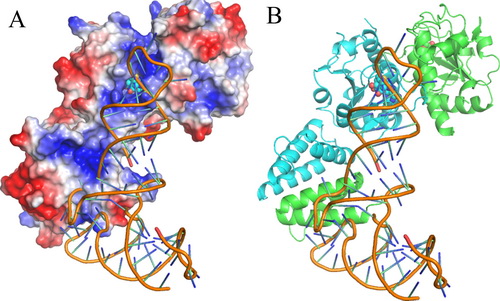
EcTrmJ与tRNA结合模式图
原文链接:
StrtRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily
原文摘要:
TrmJ proteins from the SPOUT methyltransferase superfamily aretRNA Xm32 modification enzymes that occur in bacteria and archaea. Unlike archaeal TrmJ, bacterial TrmJ require full-length tRNA moleculesas substrates. It remains unknown how bacterial TrmJs recognize substrate tRNAs and specifically catalyze a 2′-O modification at ribose 32. Herein, we demonstrate that all six Escherichia coli (Ec) tRNAs with 2′-O-methylated nucleosides at position 32 are substrates of EcTrmJ, and we show that the elbow region of tRNA, but not the amino acid acceptor stem, is needed for the methylation reaction. Our crystallographic study reveals that full-length EcTrmJ forms an unusual dimer in the asymmetric unit, with both the catalytic SPOUT domain and C-terminal extension forming separate dimeric associations. Based on these findings, we used electrophoretic mobility shift assay, isothermal titration calorimetry and enzymatic methods to identify amino acids within EcTrmJ that are involved in tRNA binding. We found that tRNA recognition by EcTrmJ involves the cooperative influences of conserved residues from both theSPOUT and extensional domains, and that this process is regulated bythe flexible hinge region that connects these two domains.
doi: 10.1093/nar/gkv745
作者:王恩多

