44个高粱重测序研究再获突破 助力粮食作物抗病育种研究
世界上最具有农业和经济价值的物种是谷类作物,主要包括小麦,大麦,玉米,水稻和高粱,全世界有超过2/3的人口以这些谷物为主粮。然而,各种病原菌的存在对粮食作物的生产造成了巨大的挑战,这也严重影响到了人类的粮食安全。世界上30%以上粮食产量的损失是由于病原菌的侵害造成的,尤其在发展中国家,病原菌爆发对粮食产量的影响更加显著。
对于减少病原菌影响,最有效可行的策略就是种植含有抗病遗传背景的品种;因此,几乎在所有的植物育种程序中,对抗病性的选择都是一个重要的组成部分。在所有的抗病基因中,核苷酸结合位点加上亮氨酸重复基因(NBS-LRR),是植物中最大,最广泛,最古老的基因家族。这些基因与各种病原菌的识别、应答有关,包括细菌,病毒,真菌,线虫,昆虫和卵菌。所以,研究抗病基因(NBS gene)对提高粮食产量,解决人类的粮食安全问题具有非常大的促进意义。
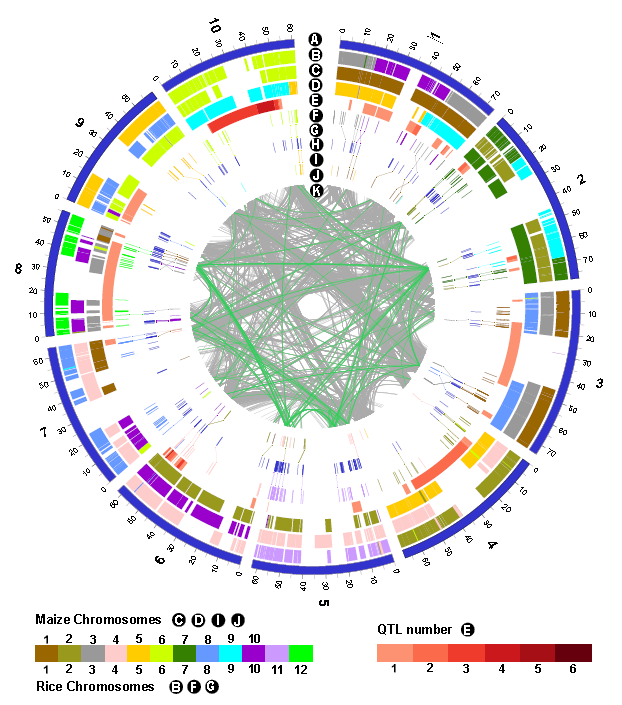
本研究中,研究人员利用同源预测的方法在高粱基因组中鉴定了346个抗病基因(NBS GENE),以多态性统计参数(θπ,θw)为基准发现抗病基因的多态性要显著高于其他基因,其中38个基因受到纯化选择,23个基因受到平衡选择,这样的发现为古老的祖先基因趋向于富集在基因组受到选择的区域提供了新的证据。同时发现,NBS编码基因也显著富集在真菌抗病QTL的区域,该类基因的多态性受到生物胁迫抗病QTL类型的影响。此外,研究人员以高粱基因组为参考绘制了高粱、玉米、水稻的共线性区域图谱,为鉴定这三个物种的同源基因提供了有力的帮助。
在此之前,大部分基因组重测序研究都是基于整个基因组的水平描述各种变异的结果,包括SNP, Indel, CNV, PAV等等,而44个高粱重测序的第二篇研究发现则利用群体遗传学的方法,对抗病基因的分子进化历程进行了非常深入的分析,得出抗病基因的适应性是由多个进化过程驱动的结论,为重测序研究的分析与阐释提供了分子进化角度的全新视角。
该项目的第一篇主文章于2013年8月28日发表在Nature Communications杂志,主要对高粱复杂的群体结构和改良驯化事件进行了深入的研究。
此外,该项目的后续分析仍在继续开展。随着大规模重测序数据的产出与发表,本次项目负责人太帅帅表示,“希望通过对高粱重测序数据的多角度深入分析,为重测序数据的分析与发表提供一个完整的范例;通过对生物学问题的深入探讨,为育种家提供有用的遗传信息,加速分子育种的进程。”
原文摘要:
The plasticity of NBS resistance genes in sorghum is driven by multiple evolutionary processes
Emma MAce, Shuaishuai Tai, David Innes, Ian Godwin, Wushu Hu, Bradley Campbell, Edward Gilding, Alan Cruickshank, Peter Prentis, Jun Wang and David Jordan
Background
Increased disease resistance is a key target of cereal breeding programs, with disease outbreaks continuing to threaten global food production, particularly in Africa. Of the disease resistance gene families, the nucleotide-binding site plus leucine-rich repeat (NBS-LRR) family is the most prevalent and ancient and is also one of the largest gene families known in plants. The sequence diversity in NBS-encoding genes was explored in sorghum, a critical food staple in Africa, with comparisons to rice and maize and with comparisons to fungal pathogen resistance QTL.
Results
In sorghum, NBS-encoding genes had significantly higher diversity in comparison to non NBS-encoding genes and were significantly enriched in regions of the genome under purifying and balancing selection, both through domestication and improvement. Ancestral genes, pre-dating species divergence, were more abundant in regions with signatures of selection than in regions not under selection. Sorghum NBS-encoding genes were also significantly enriched in the regions of the genome containing fungal pathogen disease resistance QTL; with the diversity of the NBS-encoding genes influenced by the type of co-locating biotic stress resistance QTL.
Conclusions
NBS-encoding genes are under strong selection pressure in sorghum, through the contrasting evolutionary processes of purifying and balancing selection. Such contrasting evolutionary processes have impacted ancestral genes more than species-specific genes. Fungal disease resistance hot-spots in the genome, with resistance against multiple pathogens, provides further insight into the mechanisms that cereals use in the “arms race” with rapidly evolving pathogens in addition to providing plant breeders with selection targets for fast-tracking the development of high performing varieties with more durable pathogen resistance.
作者:华大基因

