CRISPR/Cas9介导的非同源DNA敲入在多拷贝基因敲除中的应用 | BMC Biology
论文标题:Homology-independent multiallelic disruption via CRISPR/Cas9-based knock-in yields distinct functional outcomes in human cells
期刊:BMC Biology
作者:Chenzi Zhang†, Xiangjun He†, Yvonne K. Kwok, Feng Wang, Junyi Xue, Hui Zhao, Kin Wah Suen, Chi Chiu Wang, Jianwei Ren, George G. Chen, Paul B. S. Lai, Jiangchao Li, Yin Xia, Andrew M. Chan, Wai-Yee Chan and Bo Feng
发表时间:2018/12/28
数字识别码:10.1186/s12915-018-0616-2
微信链接:https://mp.weixin.qq.com/s/_G1shHW7FxO7um8QDwSHZQ
生物医学研究中,体外培养的各种细胞系是研究人类基因功能的重要模型。但是,这些细胞系在筛选建立和适应培养条件的过程中经常积累以至携带了大量的基因组异常,许多基因均呈现为三个拷贝甚至更多拷贝(图1)。因此,在研究过程中,对体外细胞系中多拷贝基因进行基因编辑并完全敲除,经常存在一定难度。

图1. LO2 细胞的双色荧光原位杂交 (Fluorescence in situ hybridization, FISH) 分析结果
原文 Figure. S1, Zhang et al. BMC Biology, 2018
之前的研究发现,由CRISPR/Cas9诱导的、经非同源末端连接路径(non-homology end joining, NHEJ pathway)介导的DNA修复,可以用于大片段DNA(或基因)在基因组中的定点敲入。 该方法的敲入效率远远高于传统的、由同源依赖性DNA修复路径(homology-dependent repair, HDR pathway)介导的敲入(He et al., Nucleic Acids Research 2016; Suzuki et al., Nature 2016; Zhou et al., FEBS Letters 2016)。因此,该方法或可为多拷贝基因敲除提供了一个很好的工具。
最近,香港中文大学生物医学学院的冯波博士课题组(张辰子等)在BMC Biology发表的最新研究工作,以LO2人类细胞系为模型,运用CRISPR/Cas9- NHEJ介导的方法, 针对目的基因同时插入了两个不携带启动子的荧光报告基因(ires-GFP/ires-Tdomato)。这些报告基因只有在以预先设计的方式插入目的基因后,才可通过目的基因启动子来表达报告荧光蛋白。随后,通过流式细胞荧光分选技术(Fluorescence activated cell sorting, FACS)对样品进行分析,可以检测并分选获得双阳性荧光细胞,以此,可一步达到同时阻断两个或者多个拷贝基因表达的目的。运用这个方法,他们对LO2中的候选基因ULK1(4拷贝),FAT10(3拷贝)进行基因敲除实验及检测(图2)。
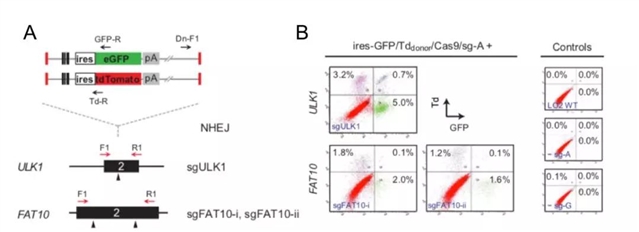
图2. 经CRIPSR/Cas9- NHEJ介导,在ULK1和FAT10基因中同时插入双荧光报告基因进行目的基因敲除
原文Figure. 2A;Zhang et al. BMC Biology, 2018
在单克隆细胞系的分析中,基因组PCR验证了在ULK1和FAT10基因的剪切位点均插入了相应荧光报告基因;mRNA、蛋白水平及功能性分析均验证了ULK1和FAT10蛋白在单克隆细胞系中的缺失(以ULK1单克隆细胞为例,图3)。而且,总体实验结果证明,该方法确实可以大幅提高获得携带目的基因敲除的单克隆细胞系的效率(原文Figure. 2D)。

图3. 对ULK1敲除单克隆细胞系的分析
原文Figure. 2E~G;Zhang et al. BMC Biology, 2018
与此同时,该研究还运用同一方法对CTIP基因(2拷贝;图1)外显子进行了双荧光报告基因的插入,并且意外地发现,在所获得的单克隆细胞系中,虽然基因组水平的PCR分析上已明确地检测到了CTIP双等位基因中均插入了荧光报告基因,但在mRNA和蛋白水平仍然可以检测到CTIP转录和蛋白的表达。
进一步对这些克隆细胞系中CTIP mRNA进行测序的结果显示,这些mRNA转录本确实来自报告基因插入后的基因组,而且携带了各种不同的剪切印记。但是意外的是,所有这些异常CTIP mRNA转录本均都保留了原有CTIP基因的阅读框架(in-frame aberrant transcripts),(图4)。

图4. CTIP敲除单克隆细胞系 (RT-PCR)测序分析结果
原文Figure. 3E, F;Zhang et al. BMC Biology, 2018
结论和意义
本研究利用CRISPR/Cas9诱导的NHEJ修复方式插入双荧光报告基因从而实现了体外细胞系中多拷贝基因的敲除。该方法可利用双荧光流式分选来对多拷贝基因敲除细胞进行富集,简化和提高了多拷贝基因敲除的效率。
另外,本研究通过对CTIP的基因编辑,发现细胞中所存在的多种DNA修复及RNA剪切机制会产生各种不同的随机产物,这使得在生存压力下,细胞将有机会富集具有功能的基因产物,使得对细胞生存起关键作用的必要基因免于被完全“敲除”。
这项研究进一步加深了对基因编辑过程中细胞反应的理解,同时,也对今后CRISPR/Cas9基因编辑的应用提供了重要的指导意义。
摘要:
Background
Cultured human cells are pivotal models to study human gene functions, but introducing complete loss of function in diploid or aneuploid cells has been a challenge. The recently developed CRISPR/Cas9-mediated homology-independent knock-in approach permits targeted insertion of large DNA at high efficiency, providing a tool for insertional disruption of a selected gene. Pioneer studies have showed promising results, but the current methodology is still suboptimal and functional outcomes have not been well examined. Taking advantage of the promoterless fluorescence reporter systems established in our previous study, here, we further investigated potentials of this new insertional gene disruption approach and examined its functional outcomes.
Results
Exemplified by using hyperploid LO2 cells, we demonstrated that simultaneous knock-in of dual fluorescence reporters through CRISPR/Cas9-induced homology-independent DNA repair permitted one-step generation of cells carrying complete disruption of target genes at multiple alleles. Through knocking-in at coding exons, we generated stable single-cell clones carrying complete disruption of ULK1 gene at all four alleles, lacking intact FAT10 in all three alleles, or devoid of intact CtIP at both alleles. We have confirmed the depletion of ULK1 and FAT10 transcripts as well as corresponding proteins in the obtained cell clones. Moreover, consistent with previous reports, we observed impaired mitophagy in ULK1−/− cells and attenuated cytokine-induced cell death in FAT10−/− clones. However, our analysis showed that single-cell clones carrying complete disruption of CtIP gene at both alleles preserved in-frame aberrant CtIPtranscripts and produced proteins. Strikingly, the CtIP-disrupted clones raised through another two distinct targeting strategies also produced varied but in-frame aberrant CtIPtranscripts. Sequencing analysis suggested that diverse DNA processing and alternative RNA splicing were involved in generating these in-frame aberrant CtIP transcripts, and some infrequent events were biasedly enriched among the CtIP-disrupted cell clones.
Conclusion
Multiallelic gene disruption could be readily introduced through CRISPR/Cas9-induced homology-independent knock-in of dual fluorescence reporters followed by direct tracing and cell isolation. Robust cellular mechanisms exist to spare essential genes from loss-of-function modifications, by generating partially functional transcripts through diverse DNA and RNA processing mechanisms.
阅读论文全文请访问:
期刊介绍:
BMC Biology(https://bmcbiol.biomedcentral.com/, 5.770 - 2-year Impact Factor, 7.556 - 5-year Impact Factor) is an open access journal publishing outstanding research in all areas of biology, with a publication policy that combines selection for broad interest and importance with a commitment to serving authors well.
(来源:科学网)

