Nature子刊:澳大利亚新研发一个可快速通过古DNA测序分析骨头小
考古和古生物挖掘坑中常常见到小碎片的骨头,不但难以识别,而且单一样本的“古DNA”(aDNA)分析也会是昂贵的和费时的。现在,澳大利亚古DNA实验室的研究人员新研发一个通过aDNA测序来分析大块骨头样本内单一不明碎片的快速方法。相关文章发表于2013年11月28日的《Scientific Reports》杂志上。
Nature子刊:澳大利亚新研发一个可快速通过古DNA测序分析骨头小碎片方法
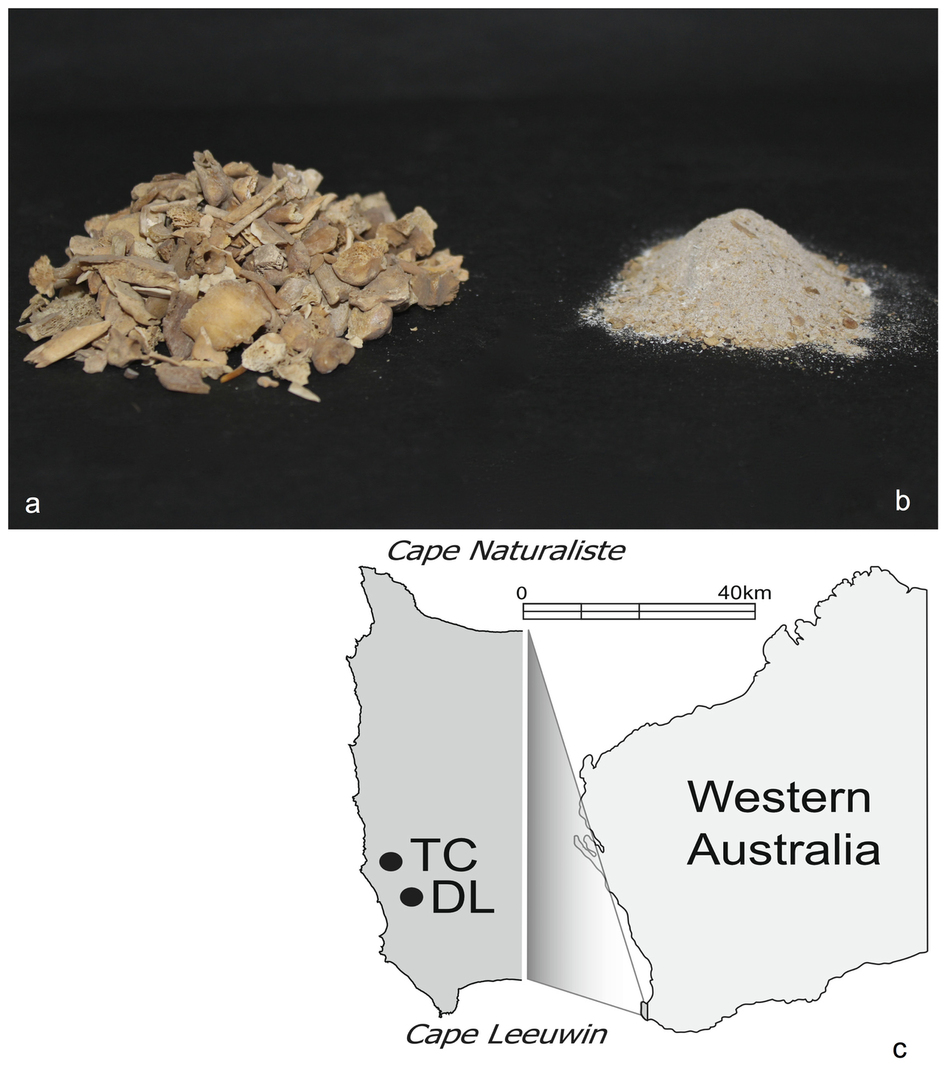
Michael Bunce及同事提出一个被称为“bulk-bone”的方法。采用该方法,他们从骨头粉末的混合样本中提取和放大时间远之距今大约46,000年前的aDNA。每个样本含有来自在澳大利亚两个考古点(其名称分别为Devil’s Lair 和 Tunnel Cave)的15个沉积层之一内所发现的50-150个化石碎片的材料。
他们对提取的aDNA进行筛选,以寻找可放大的线粒体DNA,后者随后采用高吞吐量测序方法(一种“元条码”方法)进行测序。然后,作者完成了一项分类分析,它表明这些样本含有来自一系列哺乳动物、鸟类和爬行类的DNA。
对来自发掘点和测试坑的大块骨头样本进行快速的、高性价比的分析,同时又不会损坏在形态上重要的样本,这种方法对考古学家和古生物学家来说是一个有价值的工具。
这些结果对给这种分析带来希望的一个方法做了初步演示,并且有可能帮助从原本会被扔掉的样本中揭示有关过去生物多样性的信息。然而,还需要进一步的工作,来可靠地逐步推断有关遗传多样性的信息, 并将类别鉴定提高到物种水平。
原文摘要:
Scrapheap Challenge: A novel bulk-bone metabarcoding method to investigate ancient DNA in faunal assemblages
Dáithí C. Murray, James Haile, Joe Dortch, Nicole E. White, Dalal Haouchar, Matthew I. Bellgard, Richard J. Allcock, Gavin J. Prideaux & Michael Bunce
Highly fragmented and morphologically indistinct fossil bone is common in archaeological and paleontological deposits but unfortunately it is of little use in compiling faunal assemblages. The development of a cost-effective methodology to taxonomically identify bulk bone is therefore a key challenge. Here, an ancient DNA methodology using high-throughput sequencing is developed to survey and analyse thousands of archaeological bones from southwest Australia. Fossils were collectively ground together depending on which of fifteen stratigraphical layers they were excavated from. By generating fifteen synthetic blends of bulk bone powder, each corresponding to a chronologically distinct layer, samples could be collectively analysed in an efficient manner. A diverse range of taxa, including endemic, extirpated and hitherto unrecorded taxa, dating back to c.46,000 years BP was characterized. The method is a novel, cost-effective use for unidentifiable bone fragments and a powerful molecular tool for surveying fossils that otherwise end up on the taxonomic “scrapheap”.

