Plant Journal:福建农大王凯课题组发表植物着丝粒研究成果
2016年8月24日,国际植物学权威期刊《The plant Journal》在线发表了福建农林大学海峡联合研究院王凯教授课题组和美国著名植物演化研究科学家Jonathan Wendel团队合作的关于植物功能着丝粒DNA结构与演化研究的最新研究成果(Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton),这是继不久前该团队在该杂志发表染色质空间排布研究成果后又一次基于现有科研平台开展的研究成果在该杂志发表。硕士生韩金磊,单文波及Jonathan团队的Rick Masonbrink博士为共同第一作者,王凯教授为通讯作者。
研究报道了研究人员采用染色质免疫沉淀结合细胞遗传学技术方法,对基因组演化研究的新模式材料-雷蒙德氏棉的着丝粒进行了全面解析,揭示了该物种着丝粒的DNA组成。并通过演化分析,发现其着丝粒是一种存在多种转座序列的着丝粒演化早期阶段的“特殊”材料;同时也发现了几个具有着丝粒与核仁组织区特异插入的反转座子,这一材料的获得为揭示着丝粒特异重复序列的“祖先”-着丝粒特异反转座子的出现及演化提供了宝贵的线索和研究材料。同时,该成果也首次揭示该物种着丝粒的基因组定位与结构,为物种全基因组测序后着丝粒这一黑洞的填补和揭示奠定了基础,为其他物种相关研究提供了借鉴。
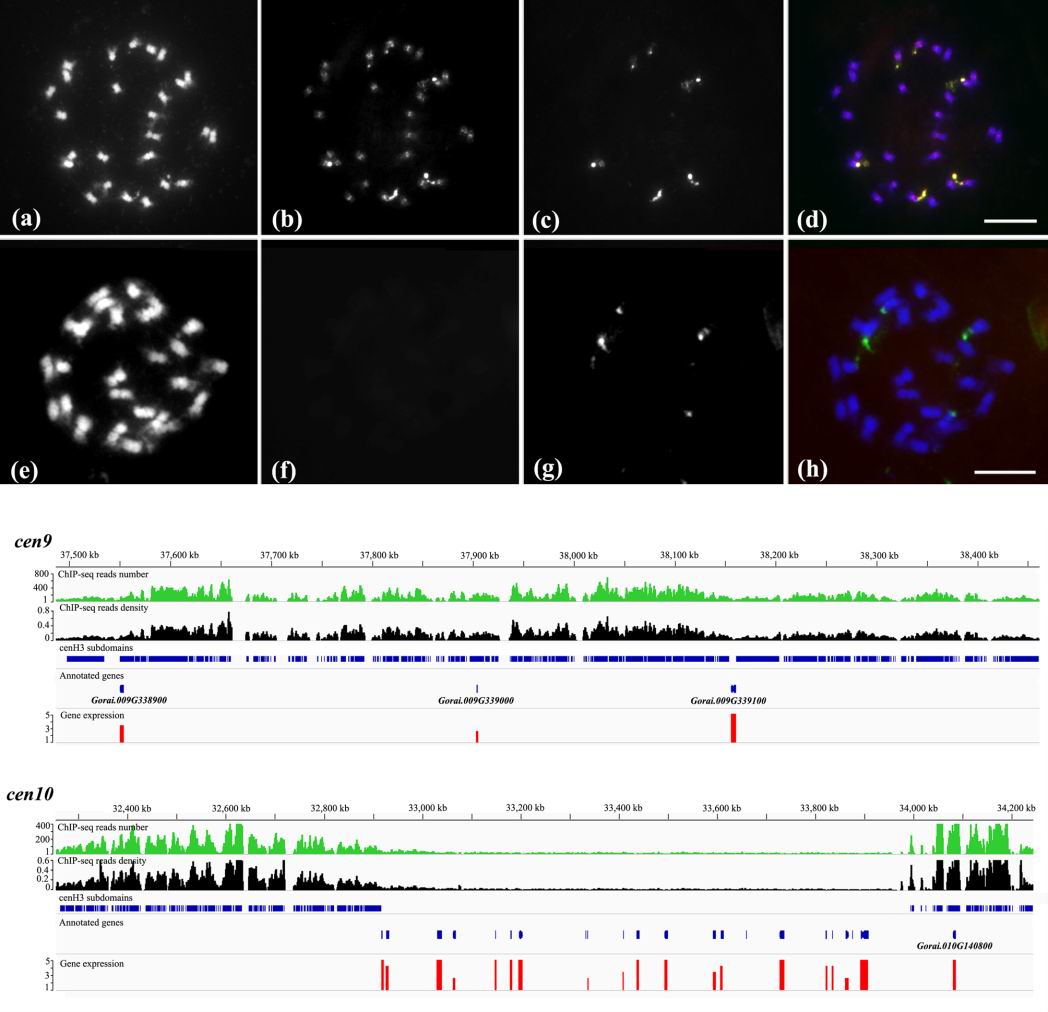
图为雷蒙德氏棉功能着丝粒结构解析
原文链接:
Rapid proLiferation and nucleolar organizer targeting centromeric retrotransposons in cotton
原文摘要:
Centromeric chromatin in most eukaryotes is composed of highly repetitive centromeric retrotransposons and satellite repeats that are highly variable even among closely related species.The evolutionary mechanisms that underlie the rapid evolution of centromeric repeatsremain unknown. To obtain insight into the evolution of centromeric repeats following polyploidy, we studied a model diploid progenitor (Gossypium raimondii,D-genome) of the allopolyploid (AD-genome) cottons, G. hirsutum and G. barbadense. Sequence analysis of chromatin-immunoprecipitated DNA showed that the G. raimondii centromeric repeats originated from retrotransposon-related sequences. Comparative analysis showed that nine of the ten analyzed centromeric repeats were absent from the centromeres in the A-genome and related diploid species (B-, F- and G-genome), indicating that they colonized the centromeres of D-genome lineage after the divergence of the A- and D- ancestral species or that they were ancestrally retained prior to the origin of Gossypium. Notably, six of the nine repeats were present in both the A- and D-subgenomes in tetraploid G. hirsutum, and increased in abundance in both subgenomes. This suggeststhat centromeric repeats may spread and proliferate between genomes subsequent to polyploidization. Two repeats, Gr334 and Gr359 occurred in both the centromeres and nucleolar organizer regions (NORs) in D and AD genome species, yet localized to just the NORs in A-, B-, F-, and G-genome species. Contained within is a story of an established centromeric repeat that is eliminated and allopolyploidization provides an opportunity for reinvasion and reestablishment, which broadens our evolutionary understanding behind the cycles of centromeric repeat establishment and targeting.
作者:王凯

